ddCRP
This is a package to apply the distance-dependent Chinese Restaurant Process (dd-CRP) to multi-dimensional graph-based data. It is based roughly on code originally written by Christopher Baldassano.
My contributions to this package are three-fold: In contrast to work presented by Baldassano et al. and Moyer et al., whose dd-CRP methods both model the univariate similarities within and between clusters, this method models the clusters themselves, placing multidimensional priors on the features of each cluster. It then explores the posterior parameter space using Collased Gibbs Sampling.
This version treats the high-dimensional feature vectors as being sampled from multivariate Gaussian distributions. In contrast to Baldassano et al. who sample similarities between feature vectors from a univariate Gaussian, and Moyer et al. who sample counts from a Poisson, this method models the full-dimensional data points themselves.
The prior distribution on features are based on an abstract class contained in
PriorsBase.pycalledPriorwith the following 3 abstract methods:Prior.sufficient_statisticsPrior.posterior_parametersPrior.marginal_evidence
Any model that implements the above three methods can be incorporated into
Priors.pyalongside the Normal-Inverse-Chi-Squared (NIX2) and Normal-Inverse-Wishart (NIW) models.On a more aesthetic level, I have refactored much of Balassano’s original Python code to make this version object-oriented and more user-friendly under the hood.
How to install and use:
git clone https://github.com/kristianeschenburg/ddCRP.git
cd ./ddCRP
pip install .
Example application to synthetic data:
We begin by importing the necessary modules. Specifically, ddCRP contains the actual algorithm, Priors contain the prior distribution models, and synthetic contains a suite of methods for generating synthetic data.
from ddCRP import ddCRP
from ddCRP import Priors
from ddCRP import synthetic
Next, we define the hyperparameters, and instantiate our prior model with these parameters. In this case, we are using the Normal-Inverse-Chi-Squared model.
alpha=10
mu = 0
kappa = 0.0001
nu = 1
sigma = 1
nix = Priors.NIX2(mu0=mu, kappa0=kappa, nu0=nu, sigma0=sigma)
Next, we sample 5-dimensional synthetic data from the same prior distribution. The synth object computes an adjacency list of our synthetic data (synth.adj_list), as well as a ground-truth label map (synth.z_).
# dimensionality of data
d = 5
# sample synthetic features for each label
# If you want to sample synthetic data from a different
# Normal-Inverse-Chi-Squared distribution,
# change kappa, mu, nu, and sigma
synth = synthetic.SampleSynthetic(kind='ell', d=d, mu0=mu, kappa0=kappa, nu0=nu, sigma0=sigma)
synth.fit()
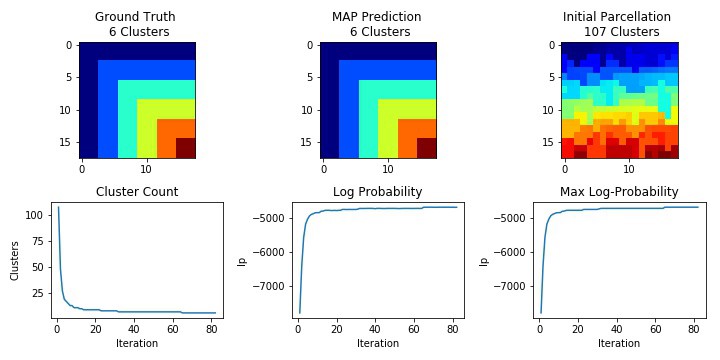
Finally, we instantiate our ddCRP model with the concentration parameter alpha, our prior model nix, along with some other parameters that govern the number of MCMC passes to perform and how often to sample diagnostic statistics about our model performance. You can see
# fit the ddCRP model
# once fitted, crp.map_z_ is the MAP label
crp = ddCRP.ddCRP(alpha, model=nix, mcmc_passes=30, stats_interval=200)
crp.fit(synth.features_, synth.adj_list, gt_z=synth.z_)
For more information on the Chinese Restaurant Process, see:
Baldassano et al. (2015), Parcellating Connectivity In Spatial Maps. PeerJ 3:e784; DOI 10.7717/peerj.784
Moyer et al. (2017), A Restaurant Process Mixture Model for Connectivity Based Parcellation of the Cortex. arXiv:1703.00981
Blei, David M. et al (2010), The Nested Chinese Restaurant Process and Bayesian Nonparametric Inference of Topic Hierarchies. JACM.