The Delta Method
Here, we’ll look at various applications of the Delta Method, especially in the context of variance stabilizing transformations, along with looking at the confidence intervals of estimates.
The Delta Method is used as a way to approximate the Standard Error of transformations of random variables, and is based on a Taylor Series approximation.
In the univariate case, if we have a random variable,
However, we don’t know the asymptotic variance of this transformed variable just yet. In this case, we can approximate our function
where
By
Slutsky’s Theorem and the
Continious Mapping Theorem, we know that since
Plugging this back in to our original equation and applying Slutsky’s Perturbation Theorem, we have:
and since we know that
The Delta Method can be generalized to the multivariate case, where, instead of the derivative, we use the gradient vector of our function:
Below, I’m going to look at a few examples applying the Delta Method to simple functions of random variables. Then I’ll go into more involved examples applying the Delta Method via
Variance Stabilizing Transformations. Oftentimes, the variance of an estimate depends on its mean, which can vary with the sample size. In this case, we’d like to find a function
We’ll start by importing the necessary libraries and defining two functions:
%matplotlib inline
import matplotlib.pyplot as plt
from matplotlib import rc
rc('text', usetex=True)
from scipy.stats import norm, poisson, expon
import numpy as np
Here, we define two simple functions – one to compute the difference between our estimate and its population paramter, and the other to compute the function of our random variable as described by the Central Limit Theorem.
def conv_prob(n, est, pop):
"""
Method to compute the estimate for convergence in probability.
"""
return (est-pop)
def clt(n, est, pop):
"""
Method to examine the Central Limit Theorem.
"""
return np.sqrt(n)*(est-pop)
Let’s have a look at an easy example with the Normal Distribution. We’ll set Scipy Normal distribution, the norm class accepts the standard deviation, not the variance. We’ll show via the Central Limit Theorem that the function
# set sample sample sizes, and number of sampling iterations
N = [5,10,50,100,500,1000]
iters = 500
mu = 0; sigma = np.sqrt(5)
# store estimates
norm_clt = {n: [] for n in N}
samples = norm(mu,sigma).rvs(size=(iters,1000))
for n in N:
for i in np.arange(iters):
est_norm = np.mean(samples[i,0:n])
norm_clt[n].append(clt(n, est_norm, mu))
Now let’s plot the results.
# Plot results using violin plots
fig = plt.subplots(figsize=(8,5))
for i,n in enumerate(N):
temp = norm_clt[n]
m = np.mean(temp)
v = np.var(temp)
print('Sample Size: %i has empirical variance: %.2f' % (n, v.mean()))
plt.violinplot(norm_clt[n], positions=[i],)

As expected, we see that the Normal distribution mean and variance estimates are independent of the sample size. In this case, we don’t need to apply a variance stabiliing transformation. We also see that the variance fluctuates around
# set sample sample sizes, and number of sampling iterations
mus = [1,2,3,4]
N = [5,10,50,100,500,1000]
iters = 2000
sigma = np.sqrt(5)
fig, ([ax1,ax2], [ax3,ax4]) = plt.subplots(2,2, figsize=(14,9))
for j ,m in enumerate(mus):
# store estimates
norm_clt = {n: [] for n in N}
samples = norm(m, sigma).rvs(size=(iters, 1000))
plt.subplot(2,2,j+1)
for k, n in enumerate(N):
np.random.shuffle(samples)
for i in np.arange(iters):
est_norm = np.mean(samples[i, 0:n])
norm_clt[n].append(clt(n, est_norm**2, m**2))
plt.violinplot(norm_clt[n], positions=[k],)

We see that the variance increases as the mean increases, and that, as the sample sizes increase, the distributions converge to the
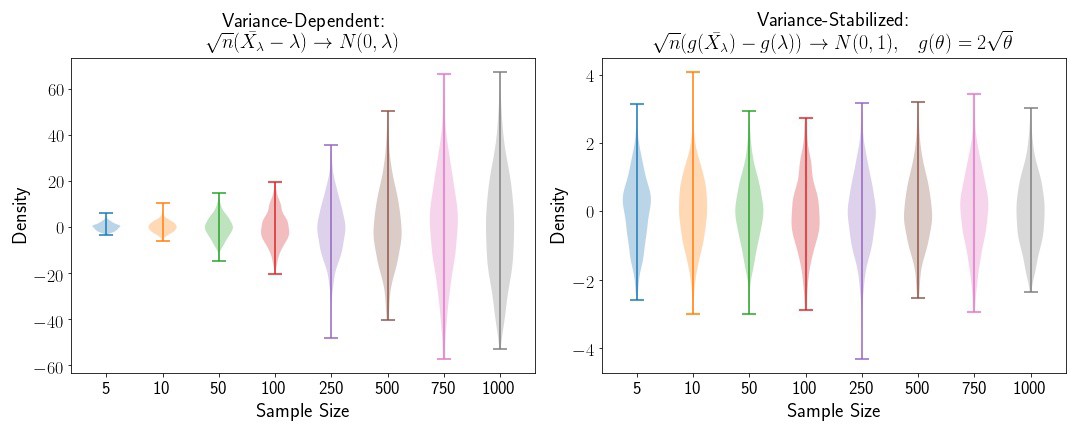
Variance Stabilization for the Poisson Distribution
Now let’s look at an example where the variance depends on the sample size. We’ll use the Poisson distribution in this case. We know that for the Poisson distribution, the variance is dependent on the mean, so let’s define a random variable,
We’ll define
If we wanted to apply the Central Limit Theorem to
where the variance
where
is our variance stabilizing function.
def p_lambda(n, theta=0.5):
"""
Function to compute lambda parameter for Poisson distribution.
Theta is constant.
"""
return n*theta
theta = 0.5
N = [5,10,50,100,250,500,750,1000]
iters = 500
clt_pois = {n: [] for n in N}
pois_novar= {n: [] for n in N}
pois_var = {n: [] for n in N}
for n in N:
for i in np.arange(iters):
est_mu = np.mean(poisson(mu=(n*theta)).rvs(n))
pois_novar[n].append(clt(n, est_mu, p_lambda(n)))
pois_var[n].append(clt(n, 2*np.sqrt(est_mu), 2*np.sqrt(p_lambda(n))))
clt_pois[n].append(conv_prob(n, est_mu, n*theta))
fig,([ax1, ax2]) = plt.subplots(2,1, figsize=(15,6))
plt.subplot(1,2,1)
for i,n in enumerate(N):
plt.violinplot(pois_novar[n], positions=[i])
plt.subplot(1,2,2)
for i,n in enumerate(N):
plt.violinplot(pois_var[n], positions=[i])
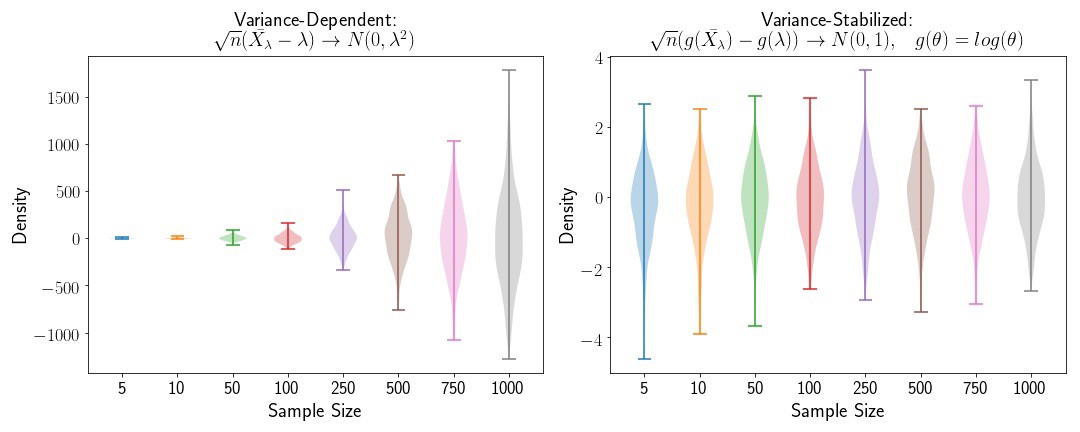
Variance Stabilization for the Exponential Distribution
Applying the same method to the Exponential distribtuion, we’ll find that the variance stabilizing transformation is
theta = 0.5
N = [5,10,50,100,250,500,750,1000]
iters = 500
clt_exp = {n: [] for n in N}
exp_novar= {n: [] for n in N}
exp_var = {n: [] for n in N}
for n in N:
for i in np.arange(iters):
samps = expon(scale=n*theta).rvs(n)
est_mu = np.mean(samps)
est_var = np.var(samps)
exp_novar[n].append(clt(n, est_mu, (n*theta)))
exp_var[n].append(clt(n, np.log(est_mu), np.log(n*theta)))
clt_exp[n].append(conv_prob(n, est_mu, n*theta))
fig,([ax1, ax2]) = plt.subplots(2,1, figsize=(15,6))
plt.subplot(1,2,1)
for i,n in enumerate(N):
plt.violinplot(exp_novar[n], positions=[i])
plt.subplot(1,2,2)
for i,n in enumerate(N):
plt.violinplot(exp_var[n], positions=[i])
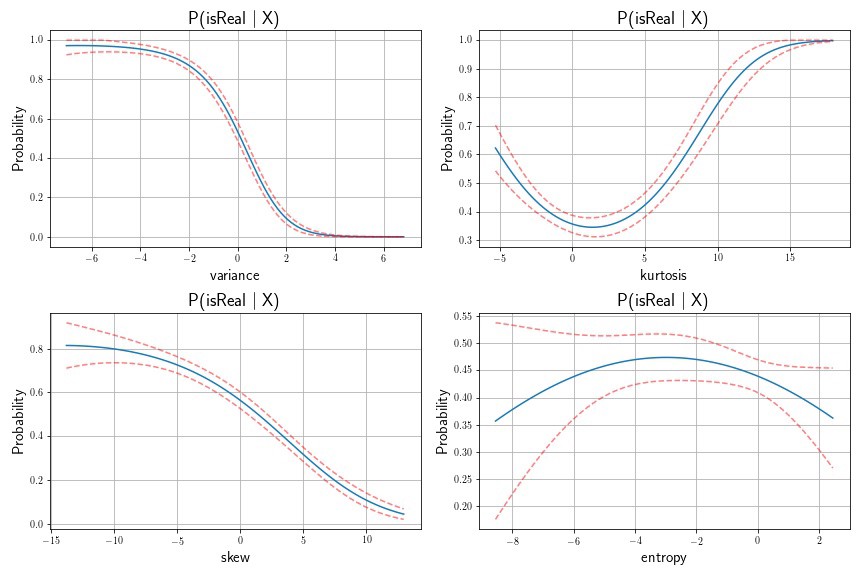
Example of Standard Error Computation Using Delta Method for Polynomial Regression
As an example of applying the Delta Method to a real-world dataset, I’ve downloaded the banknote dataset from the UCI Machine Learning Repository. In this exercise, I’ll apply the logistic function via logistic regression to assess whether or not a banknote is real or fake, using a set of features. I’ll compute confidence intervals of our prediction probabilities using the Delta Method. There are four unique predictors in this case: the variance, skew, kurtosis, and entropy of the Wavelet-transformed banknote image. I’ll treat each of these predictors independently, using polynomial basis function of degree 3.
In this example, we’re interested in the standard error of our probability estimate. Our function is the Logistic Function, as follows:
where the gradient of this multivariate function is:
so that the final estimate of our confidence interval becomes
import pandas as pd
import statsmodels.api as sm
from sklearn.preprocessing import PolynomialFeatures
import pandas as pd
import statsmodels.api as sm
from sklearn.preprocessing import PolynomialFeatures
bank = pd.read_csv('/Users/kristianeschenburg/Documents/Statistics/BankNote.txt',
sep=',',header=None, names=['variance', 'skew', 'kurtosis', 'entropy','class'])
bank.head()
fig = plt.subplots(2,2, figsize=(12,8))
for j, measure in enumerate(['variance', 'kurtosis', 'skew', 'entropy']):
predictor = np.asarray(bank[measure])
response = np.asarray(bank['class'])
idx = (response == 1)
# plot test set
plt.subplot(2,2,j+1)
plt.violinplot(predictor[idx], positions=[1]);
plt.violinplot(predictor[~idx], positions=[0])
plt.title('{:} By Classification'.format(measure), fontsize=18)
plt.ylabel('Measure: {:}'.format(measure),fontsize=15)
plt.yticks(fontsize=13)
plt.xticks([0,1],['Fake','Real'], fontsize=15)
plt.tight_layout()
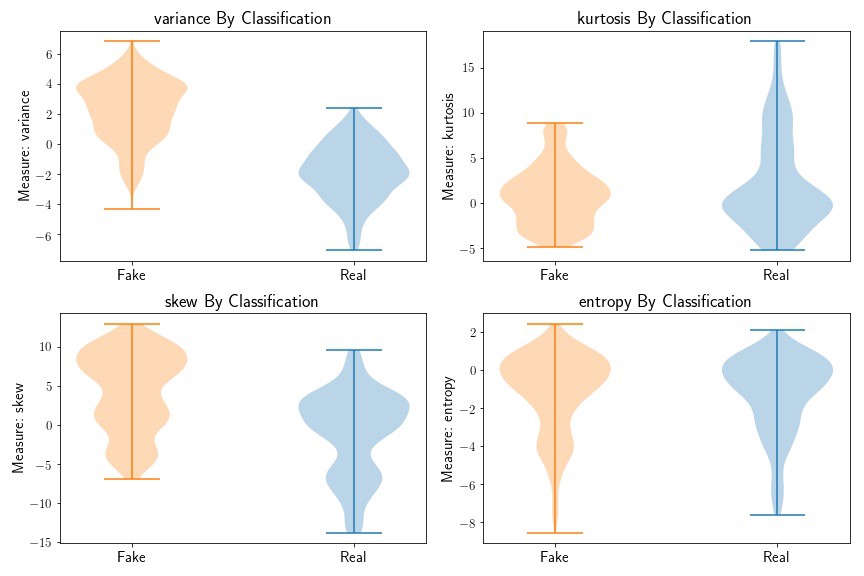
Based on the above plot, we can see that variance, skew, and kurtosis seem to be the most informative, while the entropy distributions do not seem to be that different based on bank note class.
Next, we fit a logistic regression model of note classification on note feature, with polynomial order of degree 3. We then compute the standard errors of the transformed variance. It was transformed using the logistic function, so we’ll need to compute the gradient of this function.
fig = plt.subplots(2,2, figsize=(12,8))
for j, measure in enumerate(['variance', 'kurtosis', 'skew', 'entropy']):
# Generate polynomial object to degree
# transform age to 4-degree basis function
poly = PolynomialFeatures(degree=2)
idx_order = np.argsort(bank[measure])
predictor = bank[measure][idx_order]
response = bank['class'][idx_order]
features = poly.fit_transform(predictor.values.reshape(-1,1));
# fit logit curve to curve
logit = sm.Logit(response, features).fit();
test_features = np.linspace(np.min(predictor), np.max(predictor), 100)
test_features = poly.fit_transform(test_features.reshape(-1,1))
# predict on test set
class_prob = logit.predict(test_features)
cov = logit.cov_params()
yx = (class_prob*(1-class_prob))[:,None] * test_features
se = np.sqrt(np.diag(np.dot(np.dot(yx, cov), yx.T)))
# probability can't exceed 1, or be less than 0
upper = np.maximum(0, np.minimum(1, class_prob+1.96*se))
lower = np.maximum(0, np.minimum(1, class_prob-1.96*se))
# plot test set
plt.subplot(2,2,j+1)
plt.plot(test_features[:, 1], class_prob);
plt.plot(test_features[:, 1], upper, color='red', linestyle='--', alpha=0.5);
plt.plot(test_features[:, 1], lower, color='red', linestyle='--', alpha=0.5);
plt.title(r'P(isReal \Big| X)', fontsize=18)
plt.xlabel('{:}'.format(measure),fontsize=15)
plt.ylabel('Probability',fontsize=15)
plt.grid(True)
plt.tight_layout()