Mahalanobis Distances of Brain Connectivity
For one of the projects I’m working on, I have an array of multivariate data relating to brain connectivity patterns. Briefly, each brain is represented as a surface mesh, which we represent as a graph
Additionally, for each vertex
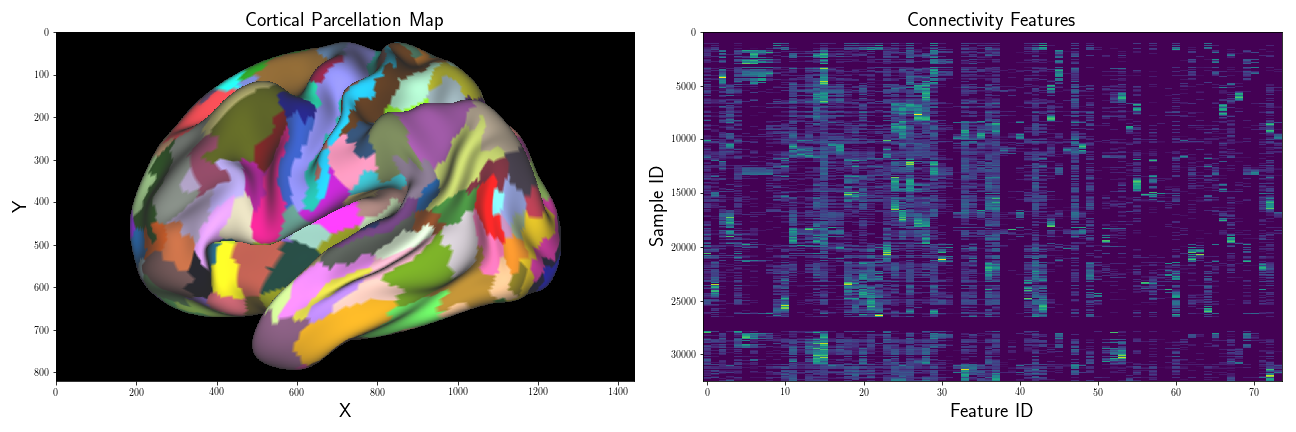
I’m interested in examining how “close” the connectivity samples of one region,
where
We know the last part is true, because the numerator and denominator are independent
where the squared Mahalanobis Distance is:
where
We know that
and consequentially, because
Therefore, we know that
so that we have
the sum of
Also, of particular importance is the fact that the Mahalanobis distance is not symmetric. That is to say, if we define the Mahalanobis distance as:
then
Now, back to the task at hand. For a specified target region,
First, I’ll estimate the covariance matrix,
Next, in order to assess whether this intra-regional similarity is actually informative, I’ll also compute the similarity of
Then, as a confirmation step to ensure that our empirical data actually follows the theoretical
See below for Python code and figures…
Step 1: Compute Parameter Estimates
%matplotlib inline
import matplotlib.pyplot as plt
from matplotlib import rc
rc('text', usetex=True)
import numpy as np
from scipy.spatial.distance import cdist
from scipy.stats import chi2, probplot
from sklearn import covariance
# lab_map is a dictionary, mapping label values to sample indices
# our region of interest has a label of 8
LT = 8
# get indices for region LT, and rest of brain
lt_indices = lab_map[LT]
rb_indices = np.concatenate([lab_map[k] for k in lab_map.keys() if k != LT])
data_lt = conn[lt_indices, :]
data_rb = conn[rb_indices, :]
# fit covariance and precision matrices
# Shrinkage factor = 0.2
cov_lt = covariance.ShrunkCovariance(assume_centered=False, shrinkage=0.2)
cov_lt.fit(data_lt)
P = cov_lt.precision_
Next, compute the Mahalanobis Distances:
# LT to LT Mahalanobis Distance
dist_lt = cdist(data_lt, data_lt.mean(0)[None,:], metric='mahalanobis', VI=P)
dist_lt2 = dist_lt**2
# fit covariance estimate for every region in cortical map
EVs = {l: covariance.ShrunkCovariance(assume_centered=False,
shrinkage=0.2) for l in labels}
for l in lab_map.keys():
EVs[l].fit(conn[lab_map[l],:])
# compute d^2 from LT to every cortical region
# save distances in dictionary
lt_to_brain = {}.fromkeys(labels)
for l in lab_map.keys():
temp_data = conn[label_map[l], :]
temp_mu = temp_data.mean(0)[None, :]
temp_mh = cdist(data_lt, temp_mu, metric='mahalanobis', VI=EVs[l].precision_)
temp_mh2 = temp_mh**2
lt_to_brain[l] = temp_mh2
# plot distributions seperate (scales differ)
fig = plt.subplots(2,1, figsize=(12,12))
plt.subplot(2,1,1)
plt.hist(lt_to_brain[LT], 50, density=True, color='blue',
label='Region-to-Self', alpha=0.7)
plt.subplot(2,1,2)
for l in labels:
if l != LT:
plt.hist(lt_to_brain[l], 50, density=True, linewidth=2,
alpha=0.4, histtype='step')
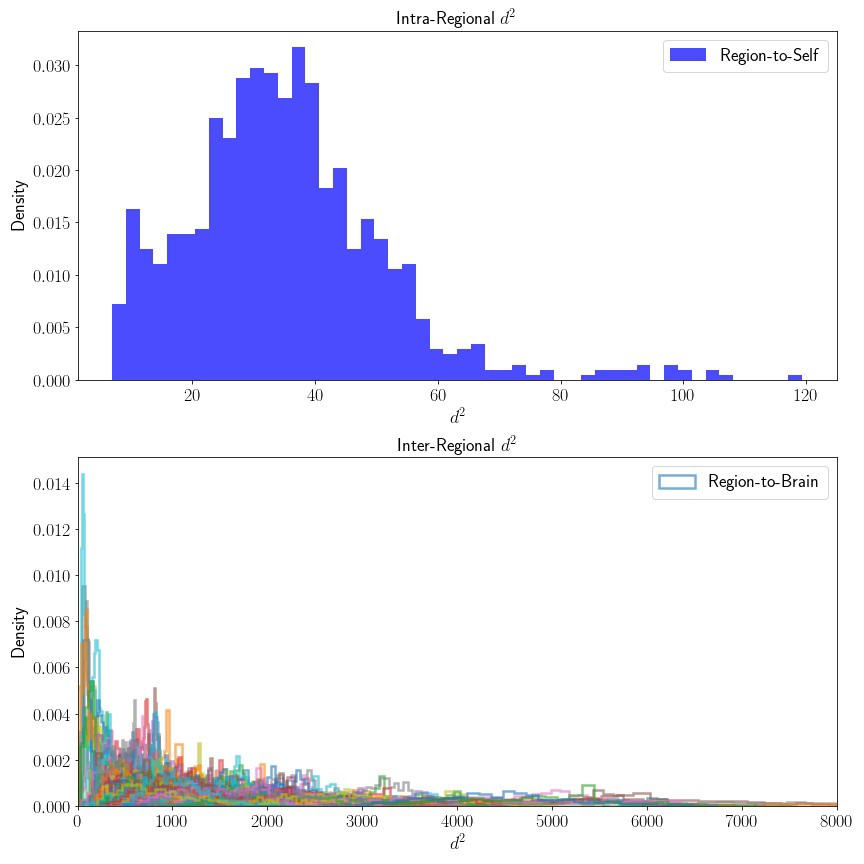
As expected, the distribution of
Step 2: Distributional QC-Check
Because we know that our data should follow a
p = data_lt.shape[1]
mle_chi2_theory = chi2.fit(dist_lt2, fdf=p)
xr = np.linspace(data_lt.min(), data_lt.max(), 1000)
pdf_chi2_theory(xr, *mle_chi2_theory)
fig = plt.subplot(1,2,2,figsize=(18, 6))
# plot theoretical vs empirical null distributon
plt.subplot(1,2,1)
plt.hist(data_lt, density=True, color='blue', alpha=0.6,
label = 'Empirical')
plt.plot(xr, pdf_chi2_theory, color='red',
label = '$\chi^{2}_{p}')
# plot QQ plot of empirical distribution
plt.subplot(1,2,2)
probplot(D2.squeeze(), sparams=mle_chi2_theory, dist=chi2, plot=plt);
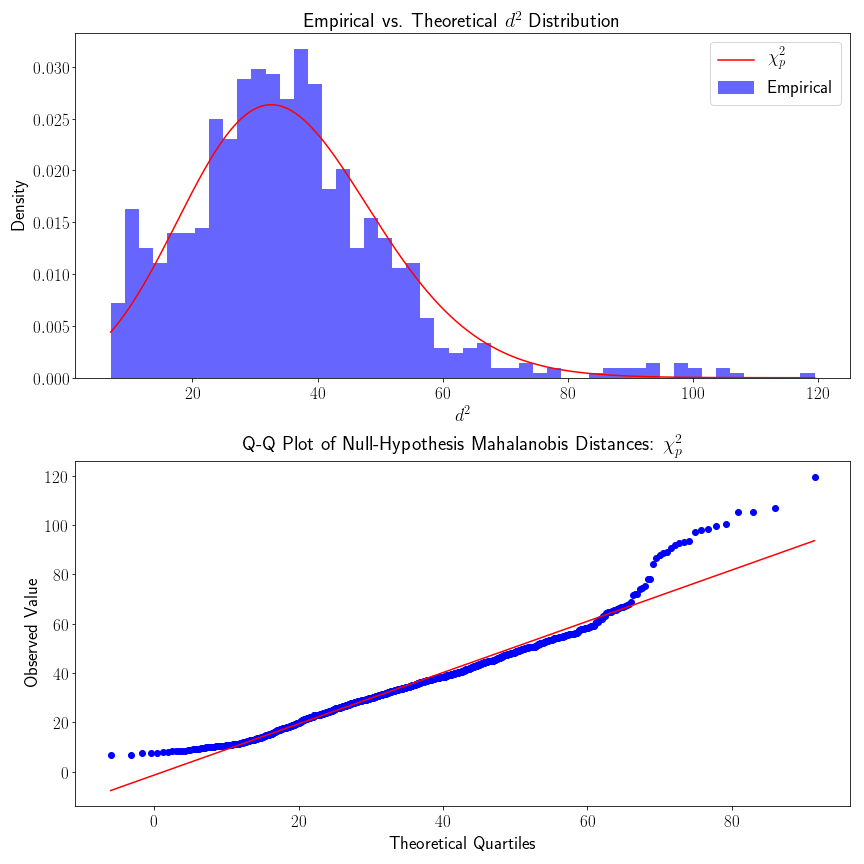
From looking at the QQ plot, we see that the empirical density fits the theoretical density pretty well, but there is some evidence that the empirical density has heavier tails. The heavier tail of the upper quantile could probability be explained by acknowledging that our starting cortical map is not perfect (in fact there is no “gold-standard” cortical map). Cortical regions do not have discrete cutoffs, although there are reasonably steep
gradients in connectivity. If we were to include samples that were considerably far away from the the rest of the samples, this would result in inflated densities of higher
Likewise, we also made the distributional assumption that our connectivity vectors were multivariate normal – this might not be true – in which case our assumption that
Finally, let’s have a look at some brains! Below, is the region we used as our target – the connectivity profiles from vertices in this region were used to compute our mean vector and covariance matrix – we compared the rest of the brain to this region.
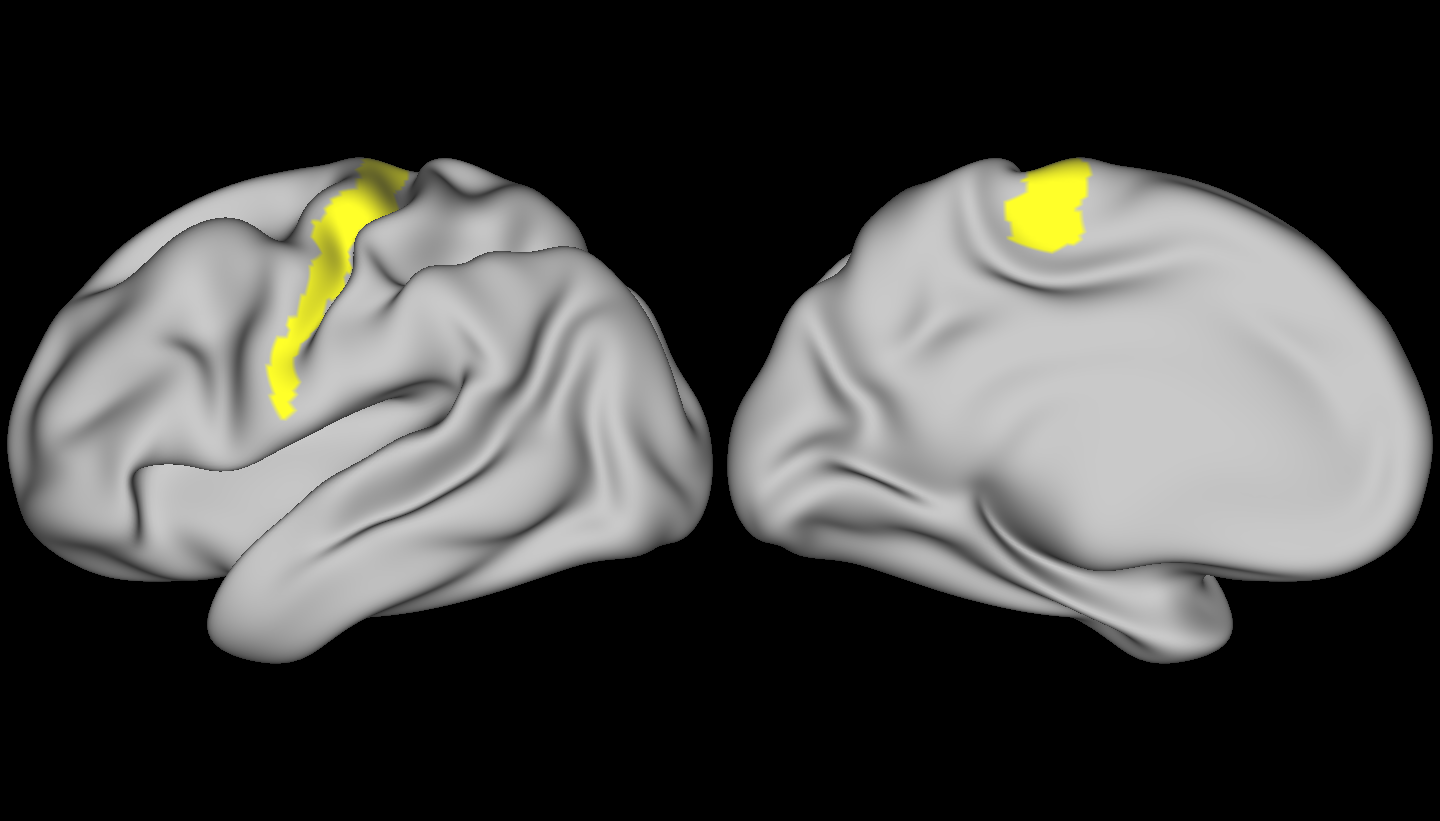
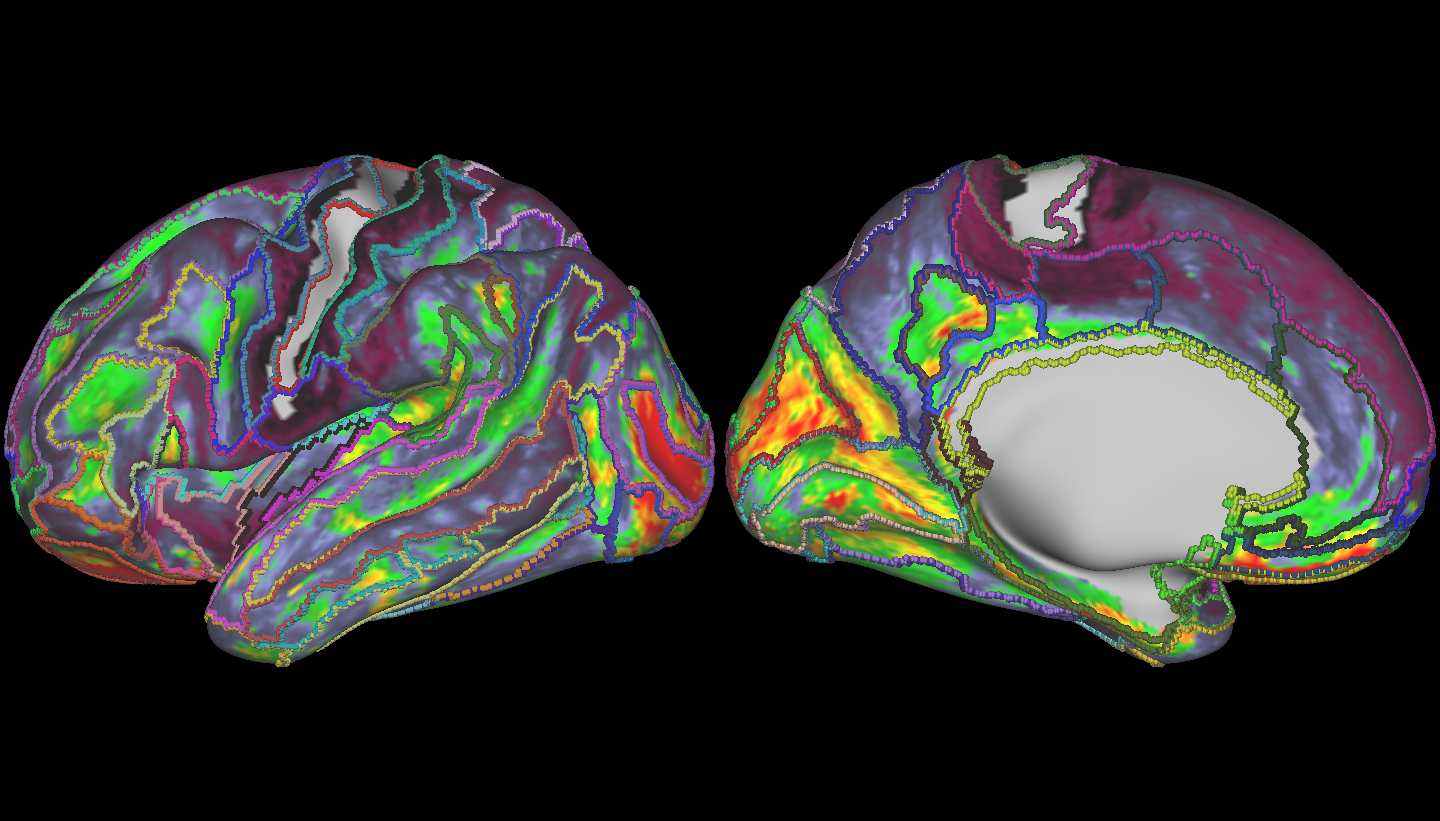
Here, larger
This is interesting stuff – I’d originally intended on just learning more about the Mahalanobis Distance as a measure, and exploring its distributional properties – but now that I see these results, I think it’s definitely worth exploring further!